All life is composed of cells several orders of magnitude smaller than a grain of salt. Their seemingly simple-looking structures hide the intricate and complex molecular activity that enables them to perform the life-sustaining functions.
Researchers are beginning to be able to visualize this activity in a level of detail they were previously unable to.
Biological structures can be visualized either by starting at the level of the whole organism and working down, or by starting at the level of individual atoms and working up.
However, there was a resolution gap between a cell’s smallest structures, like the cytoskeleton, which supports the cell’s shape, and its largest structures, like the ribosomes, which make proteins in cells.
By analogy with Google Maps, while scientists could see entire cities and individual houses, they didn’t have the tools to see how the houses came together to form neighborhoods.
Recognizing these neighborhood-level details is essential to understanding how individual components in a cell’s environment work together.
New tools are constantly closing this gap. And the further development of a certain technique, Cryo-electron tomography or cryo-EThas the potential to deepen how researchers study and understand how cells function in health and disease.
As former Editor-in-Chief of Science magazine and as researcher After decades of studying difficult-to-visualize large protein structures, I have witnessed amazing advances in the development of tools that can determine biological structures in detail.
Just as it becomes easier to understand how complicated systems work when you know what they look like, understanding how biological structures fit together inside a cell is key to understanding how organisms work.
A Brief History of Microscopy
In the 17th century, light microscope first revealed the existence of cells. In the 20th century, electron microscopy offered even more detail and revealed the sophisticated structures within the cellsincluding organelles such as the endoplasmic reticulum, a complex network of membranes that play key roles in protein synthesis and transport.
From the 1940s to the 1960s, biochemists worked to dissect cells into their molecular components and learn how to determine the 3D structures of proteins and other macromolecules at or near atomic resolution. This was initially done using X-ray crystallography to visualize the structure of myoglobina protein that supplies oxygen to the muscles.
In the last decade, techniques based on nuclear magnetic resonancewhich produces images based on how atoms interact in a magnetic field, and cryo-electron microscopy have rapidly increased the number and complexity of structures scientists can visualize.
frameborder=”0″ allow=”accelerometer; autoplay; write clipboard; encrypted media; gyroscope; picture in picture; web-share” allowfullscreen>
What is cryo-EM and cryo-ET?
Cryo-Electron Microscopy or Cryo-EMuses a camera to see how an electron beam deflects as the electrons pass through a sample to reveal structures at the molecular level.
Samples are quickly frozen to protect them from radiation damage. Detailed models of the structure of interest are created by taking multiple images of individual molecules and averaging them into a 3D structure.
cryo ET shares similar components with cryo-EM but uses different methods. Since most cells are too thick to be imaged sharply, an area of interest in a cell is first thinned out with an ion beam.
The sample is then tilted to take multiple images of it at different angles, analogous to a CT scan of a body part – although in this case the imaging system itself is tilted, not the patient. These images are then combined by a computer to create a 3D image of part of the cell.
The resolution of this image is high enough for researchers – or computer programs – to be able to identify the individual components of different structures in a cell. For example, researchers have used this approach to show how proteins move and break down inside you algal cell.
Many of the steps that researchers previously had to perform manually to determine the structures of cells are being automated, allowing scientists to identify new structures at a much faster rate.
For example, combine cryo-EM with artificial intelligence programs like AlphaFold can facilitate image interpretation by predicting protein structures that have not yet been characterized.
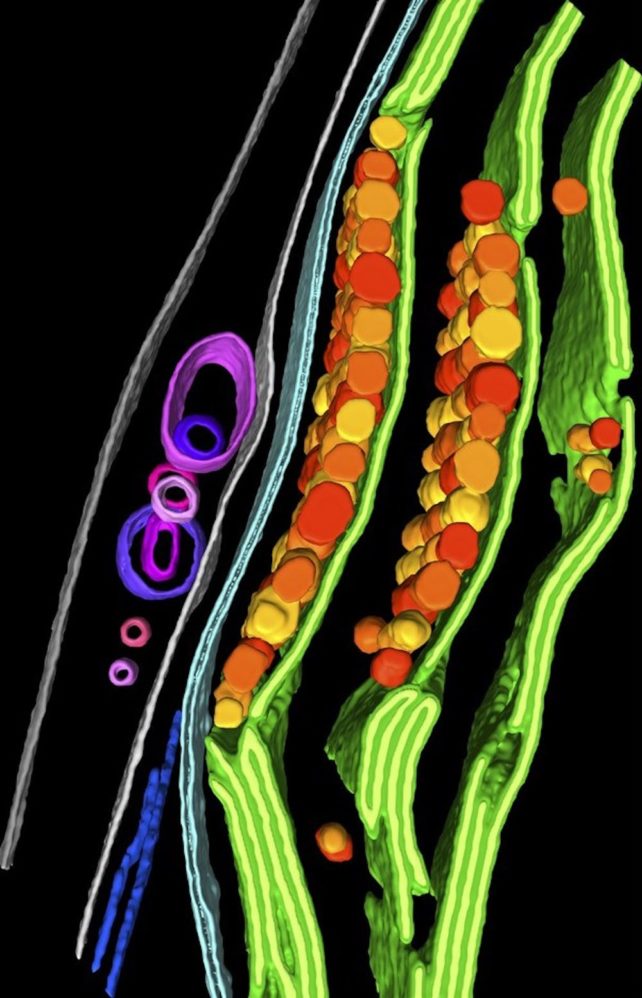
Understand cell structure and function
As imaging techniques and workflows improve, researchers will be able to address some key questions in cell biology with different strategies.
The first step is to decide which cells and which regions within those cells to study. Another visualization technique called correlated light and electron microscopy or CLEMuses fluorescent tags to localize regions where interesting processes take place in living cells.
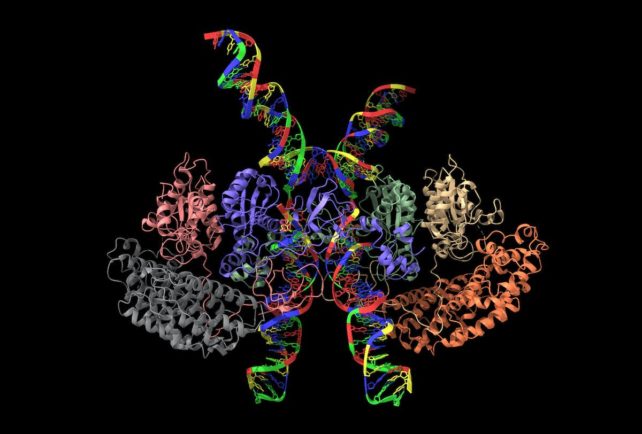
If you compare them genetic difference between cells can provide additional insights. Scientists can look at cells that are unable to perform certain functions and see how this is reflected in their structure. This approach can also help researchers study how cells interact with each other.
Cryo-ET is likely to remain a specialized tool for some time to come. But further technological developments and increasing accessibility will allow the scientific community to study the connection between cellular structure and function at previously inaccessible levels of detail.
I await new theories on how we understand cells, moving from disorganized bags of molecules to intricately organized and dynamic systems.![]()
Jeremy BergProfessor of Computational and Systems Biology, Associate Senior Vice Chancellor for Science Strategy and Planning, University of Pittsburgh
This article is republished by The conversation under a Creative Commons license. read this original article.


 Taxidermy swan one of 150 artifacts in ‘Orlando Collected’ exhibit
Taxidermy swan one of 150 artifacts in ‘Orlando Collected’ exhibit


