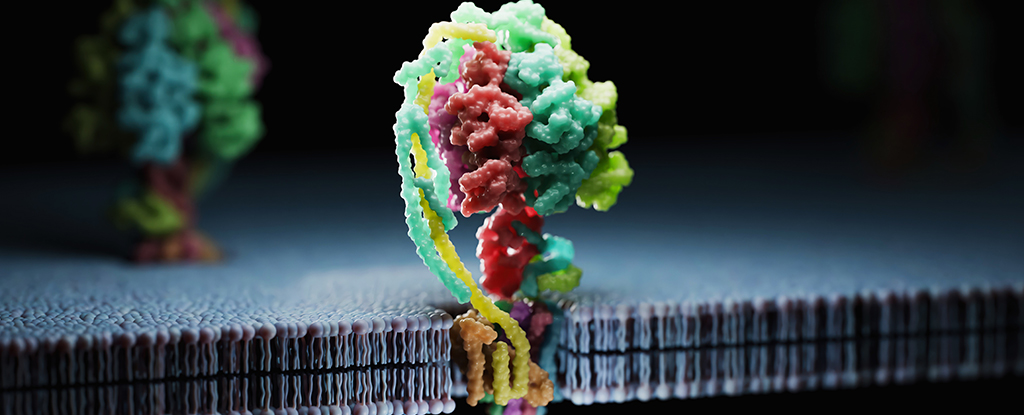
Synthesizing new proteins – the building blocks of biological life – is a scientific field of immense potential, and a newly developed AI model promises to create instructions for new proteins way beyond those found in nature.
Scientists in the US have used EvolutionaryScale Model 3 (ESM3) to synthesize a new protein called esmGFP (green fluorescent protein), which only shares 58 percent of its material with its closest natural relative tagRFP.
That’s the equivalent of 500 million years of evolution being processed by AI, the research team estimates, and it opens the way to creating custom-made proteins that can be designed for specific uses, or unlocking more functions from existing proteins.
“More than three billion years of evolution have produced an image of biology encoded into the space of natural proteins,” write the researchers, led by Thomas Hayes, founder of EvolutionaryScale in New York, in their published paper.
“Here we show that language models trained at scale on evolutionary data can generate functional proteins that are far away from known proteins.”
I’m so excited to share what we’ve been working on @EvoscaleAI. ESM3 is a multimodal generative masked language model for programming biology. Here’s a short thread on the architecture behind ESM3.
https://t.co/jldHYRAPNy
— Thomas Hayes (@THayes427) June 25, 2024
ESM3 was trained on an impressive 3.15 billion protein sequences (the order of amino acids in a protein), 236 million protein structures (their 3D shapes), and 539 million protein annotations (descriptive labels).
By spotting patterns in those vast troves of data, the AI model can understand what works and what doesn’t in protein building and function – in the same way that ChatGPT can compose a new poem that rhymes after reading millions of poems written by humans.
What makes esmGFP extra special is that it works: it’s fluorescent just like its relative tagRFP. Fluorescent proteins give some ocean organisms their glow, and their use as markers have huge importance in medicine and biotechnology.
“We chose the functionality of fluorescence because it is difficult to achieve, easy to measure, and one of the most beautiful mechanisms in nature,” the team writes.
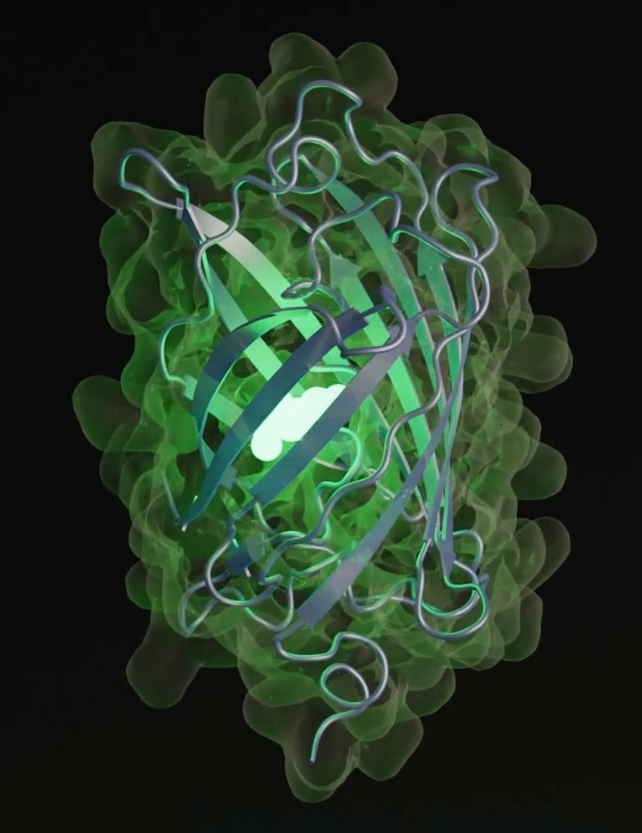
The AI takes away a lot of the trial and error in protein synthesis, while adding the ability to explore far away from proteins we currently know about.
“Proteins can be seen as existing within an organized space where each protein is neighbored by every other that is one mutational event away,” write the researchers. “The structure of evolution appears as a network within this space, connecting all proteins by the paths that evolution can take between them.”
For evolution to occur, the team says each protein must change into the next one without the system of which it is a part losing its overall functionality. A language model recognizes proteins in this space.
Proteins designed by ESM3 still need to be validated, synthesized, and tested, which takes time, but the team is confident of making further progress here. In the not-too-distant future we could be producing proteins for everything from medicines to biomaterials just with some clever AI prompting.
“Protein language models do not explicitly work within the physical constraints of evolution, but instead can implicitly construct a model of the multitude of potential paths evolution could have followed,” the researchers explain.
The research has been published in Science.